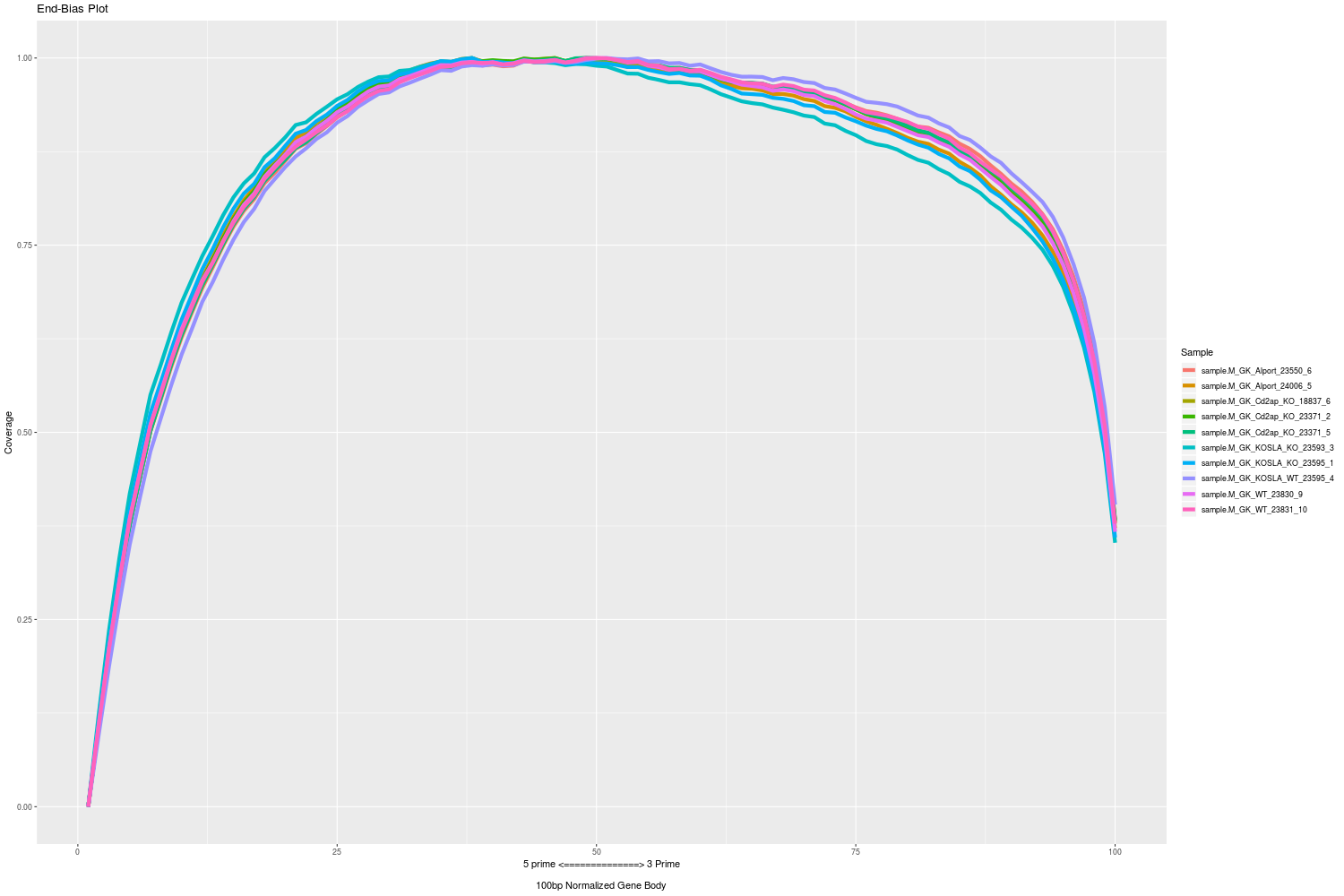
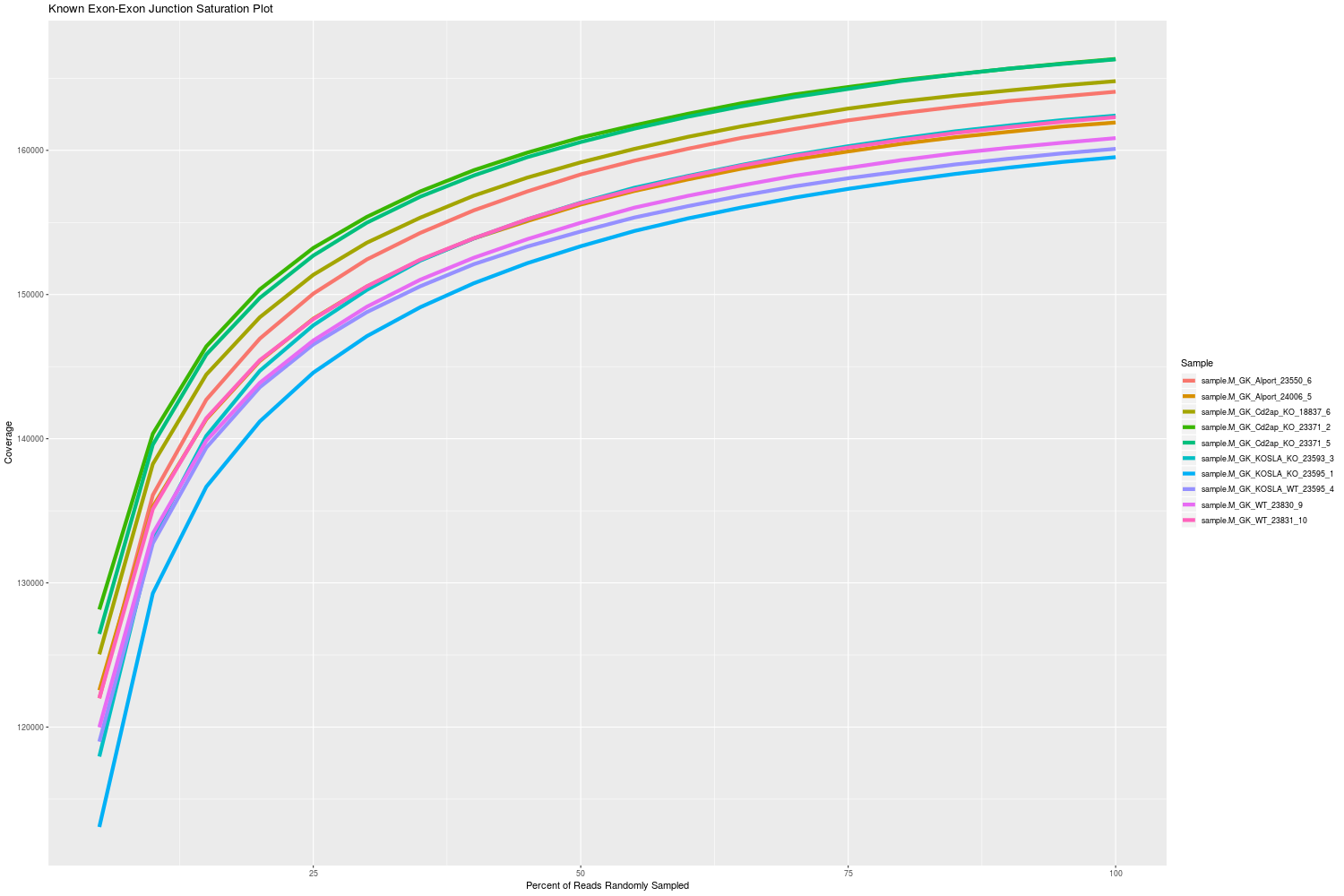
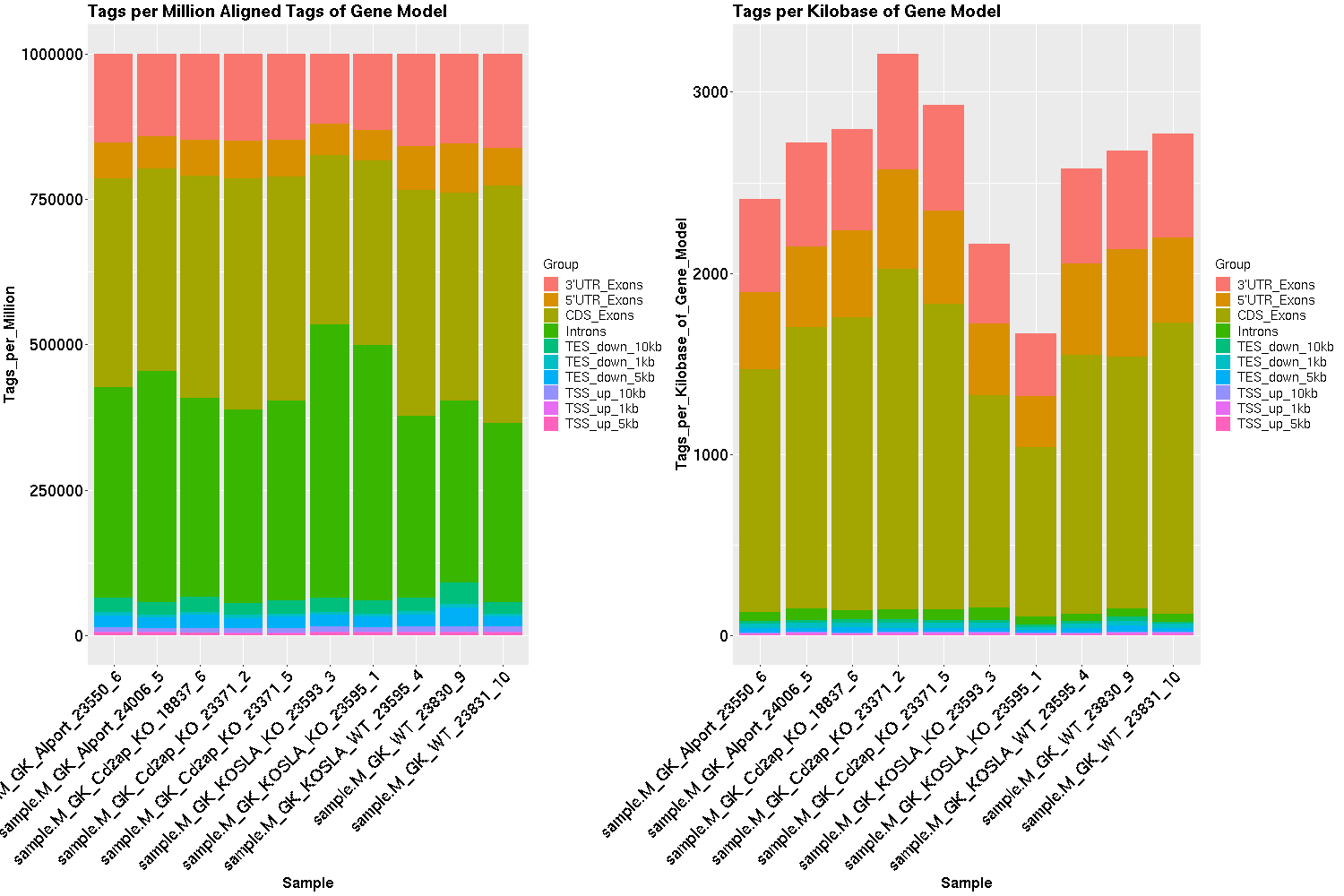
Mosammaparast Lab
HiSeq 2990_8 Project Details:
Analyzed: 06/20/2019
1 Lane(s) single reads: 1 X 50 nts
Aligned to h_sapien reference build Ensembl_R76
| Sample | Total Reads | Read Depth Quality | Mapped Reads | Percent Mapped Reads | Percent Mapped Quality | Unique Mapped Reads | Percent Unique Mapped Reads | Multi Mapped Reads | Percent Multi Mapped Reads | Percent Unmapped Too Many Loci | Percent Unmapped Too Many Mismatches | Percent Unmapped Too Short | Percent Unmapped Other | Avg Input Length | Avg Mapped Length | Spliced Reads | Annotated Spliced Reads | Spliced GT/AG | Spliced GC/AG | Spliced AT/AC | Noncanonical Spliced Reads | Mismatch Rate | Deletion Rate | Avg Deletion Length | Insertion Rate | Avg Insertion Length | Chimeric Reads | Percent Chimeric Reads |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample.M_GK_Alport_23550_6 | 64316774 | Pass | 63657705 | 98.98 | Pass | 55979136 | 87.04 | 7678569 | 11.94 | 0.38 | 0.44 | 0.01 | 0.19 | 284 | 243.2 | 20530604 | 20139876 | 20299942 | 155227 | 14906 | 60529 | 0.29 | 0.01 | 2.21 | 0.02 | 2.4 | 0 | 0 |
| sample.M_GK_Alport_24006_5 | 75008681 | Pass | 74318347 | 99.08 | Pass | 66413546 | 88.54 | 7904801 | 10.54 | 0.28 | 0.42 | 0.01 | 0.21 | 285 | 254.96 | 23897075 | 23400847 | 23626092 | 187410 | 17211 | 66362 | 0.24 | 0.01 | 2.12 | 0.01 | 2.11 | 0 | 0 |
| sample.M_GK_Cd2ap_KO_18837_6 | 67904449 | Pass | 67309975 | 99.12 | Pass | 59881240 | 88.18 | 7428735 | 10.94 | 0.23 | 0.45 | 0.01 | 0.18 | 285 | 256.97 | 25714705 | 25295620 | 25428414 | 190880 | 18057 | 77354 | 0.32 | 0.02 | 2.44 | 0.02 | 2.81 | 0 | 0 |
| sample.M_GK_Cd2ap_KO_23371_2 | 75048547 | Pass | 74496767 | 99.27 | Pass | 66702855 | 88.88 | 7793912 | 10.39 | 0.21 | 0.33 | 0.01 | 0.19 | 286 | 262.25 | 29705314 | 29213765 | 29392331 | 218847 | 20400 | 73736 | 0.24 | 0.01 | 2.13 | 0.01 | 2.34 | 0 | 0 |
| sample.M_GK_Cd2ap_KO_23371_5 | 70871004 | Pass | 70331740 | 99.24 | Pass | 62896487 | 88.75 | 7435253 | 10.49 | 0.22 | 0.35 | 0.01 | 0.18 | 285 | 257.67 | 26537926 | 26085846 | 26253395 | 194790 | 18048 | 71693 | 0.27 | 0.01 | 2.21 | 0.02 | 2.52 | 0 | 0 |
| sample.M_GK_KOSLA_KO_23593_3 | 68944235 | Pass | 68315723 | 99.09 | Pass | 61024274 | 88.51 | 7291449 | 10.58 | 0.3 | 0.33 | 0.02 | 0.27 | 284 | 257.48 | 17630882 | 17236734 | 17401161 | 134611 | 11714 | 83396 | 0.29 | 0.02 | 2.52 | 0.02 | 2.47 | 0 | 0 |
| sample.M_GK_KOSLA_KO_23595_1 | 50084006 | Pass | 49659700 | 99.15 | 2011/04/25 | 44120777 | 88.09 | 5538923 | 11.06 | 0.29 | 0.30 | 0.02 | 0.24 | 283 | 251.73 | 14216434 | 13906878 | 14026811 | 113130 | 10230 | 66263 | 0.28 | 0.02 | 2.52 | 0.02 | 2.4 | 0 | 0 |
| sample.M_GK_KOSLA_WT_23595_4 | 61868581 | Pass | 61121964 | 98.8 | Pass | 53564105 | 86.58 | 7557859 | 12.22 | 0.41 | 0.32 | 0.01 | 0.46 | 285 | 251.7 | 21775235 | 21276793 | 21276793 | 184463 | 15240 | 132079 | 0.35 | 0.02 | 0.02 | 0.02 | 2.48 | 0 | 0 |
| sample.M_GK_WT_23830_9 | 63779284 | Pass | 63102888 | 98.94 | Pass | 55187946 | 86.53 | 7914942 | 12.41 | 0.21 | 0.69 | 0.01 | 0.15 | 284 | 248.47 | 21554470 | 21106561 | 21262804 | 190360 | 15400 | 85906 | 0.49 | 0.03 | 2.92 | 0.04 | 3.27 | 0 | 0 |
| sample.M_GK_WT_23831_10 | 65021486 | Pass | 64014215 | 98.45 | Pass | 56263690 | 86.53 | 7750525 | 11.92 | 0.48 | 0.32 | 0.01 | 0.73 | 284 | 254.01 | 24268745 | 23638662 | 23861755 | 203280 | 17518 | 186192 | 0.33 | 0.02 | 2.44 | 0.02 | 2.24 | 0 | 0 |
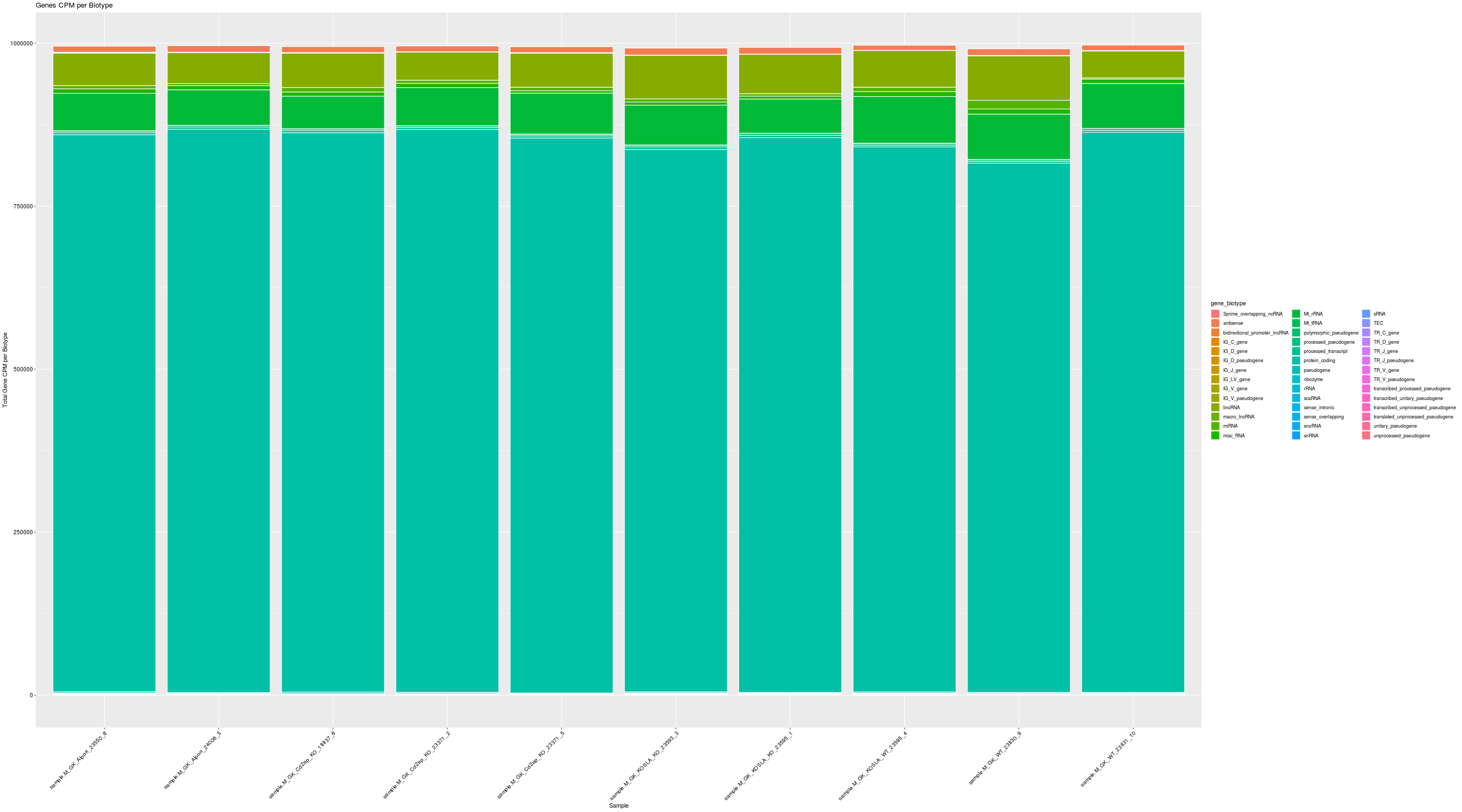
| Sample | Features.Detected | Total.Counts | Features.greater.than.5.CPM | Features.greater.than.50.CPM | Features.greater.than.100.CPM | Ribosomal.Features.Detected | Ribosomal.Total.Counts | Ribosomal.Fraction | Ribosomal.Features.greater.than.5.CPM | Ribosomal.Features.greater.than.50.CPM | Ribosomal.Features.greater.than.100.CPM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| sample.M_GK_Alport_23550_6 | 27280 | 30109192 | 12836 | 4330 | 1993 | 111 | 1727929 | 0.0573887535739916 | 26 | 4 | 4 |
| sample.M_GK_Alport_24006_5 | 26711 | 33311818 | 12752 | 4411 | 2021 | 113 | 1669809 | 0.050126624731199 | 26 | 4 | 4 |
| sample.M_GK_Cd2ap_KO_18837_6 | 26711 | 33311818 | 12752 | 4411 | 2021 | 113 | 1669809 | 0.050126624731199 | 26 | 4 | 4 |
| sample.M_GK_Cd2ap_KO_23371_2 | 26663 | 38211313 | 12608 | 4427 | 2042 | 116 | 2240148 | 0.0586252558241063 | 21 | 4 | 4 |
| sample.M_GK_Cd2ap_KO_23371_5 | 26773 | 35363992 | 12734 | 4373 | 1980 | 112 | 2220118 | 0.0627790550342846 | 21 | 4 | 4 |
| sample.M_GK_KOSLA_KO_23593_3 | 27183 | 26383889 | 13037 | 4336 | 1927 | 119 | 1612378 | 0.0611122188999506 | 37 | 4 | 4 |
| sample.M_GK_KOSLA_KO_23595_1 | 26493 | 20658777 | 12949 | 99.15 | 2055 | 44120777 | 88.09 | 0.0525303119347288 | 11.06 | 4 | 4 |
| sample.M_GK_KOSLA_WT_23595_4 | 29766 | 30999093 | 12387 | 4137 | 1945 | 132 | 2222044 | 0.0716809359551262 | 17 | 4 | 4 |
| sample.M_GK_WT_23830_9 | 26511 | 31738085 | 12342 | 4034 | 1821 | 106 | 2211153 | 0.0696687591579643 | 15 | 4 | 4 |
| sample.M_GK_WT_23831_10 | 30858 | 33268720 | 12525 | 4214 | 1960 | 122 | 2295971 | 0.0690129046143044 | 13 | 4 | 4 |
| Sample | Features.Detected | Total.Counts | Features.greater.than.5.CPM | Features.greater.than.50.CPM | Features.greater.than.100.CPM |
|---|---|---|---|---|---|
| sample.M_GK_Alport_23550_6 | 48991 | 10821987.4970489 | 17846 | 4230 | 1941 |
| sample.M_GK_Alport_24006_5 | 50070 | 13531547.8923394 | 17959 | 4253 | 2030 |
| sample.M_GK_Cd2ap_KO_18837_6 | 49666 | 13440015.2136905 | 17759 | 4313 | 1964 |
| sample.M_GK_Cd2ap_KO_23371_2 | 51245 | 17092328.2648094 | 17740 | 4271 | 1978 |
| sample.M_GK_Cd2ap_KO_23371_5 | 50667 | 14568856.8309889 | 17925 | 4214 | 1929 |
| sample.M_GK_KOSLA_KO_23593_3 | 49810 | 9860666.08229841 | 19304 | 4110 | 1806 |
| sample.M_GK_KOSLA_KO_23595_1 | 47813 | 7387891.1357028 | 18983 | 4345 | 1966 |
| sample.M_GK_KOSLA_WT_23595_4 | 48877 | 11583400.1676355 | 16803 | 4092 | 1909 |
| sample.M_GK_WT_23830_9 | 48091 | 11411271.5056075 | 17102 | 4219 | 1947 |
| sample.M_GK_WT_23831_10 | 50352 | 12844795.3415866 | 16859 | 4161 | 1925 |
| First name | Last name | Position | Office | Age | Start date | Salary | Extn. | |
|---|---|---|---|---|---|---|---|---|
| Tiger | Nixon | System Architect | Edinburgh | 61 | 2011/04/25 | $320,800 | 5421 | t.nixon@datatables.net |
| Garrett | Winters | Accountant | Tokyo | 63 | 2011/07/25 | $170,750 | 8422 | g.winters@datatables.net |
| Ashton | Cox | Junior Technical Author | San Francisco | 66 | 2009/01/12 | $86,000 | 1562 | a.cox@datatables.net |
| Cedric | Kelly | Senior Javascript Developer | Edinburgh | 22 | 2012/03/29 | $433,060 | 6224 | c.kelly@datatables.net |
| Airi | Satou | Accountant | Tokyo | 33 | 2008/11/28 | $162,700 | 5407 | a.satou@datatables.net |
| Brielle | Williamson | Integration Specialist | New York | 61 | 2012/12/02 | $372,000 | 4804 | b.williamson@datatables.net |

| First name | Last name | Position | Office | Age | Start date | Salary | Extn. | |
|---|---|---|---|---|---|---|---|---|
| Tiger | Nixon | System Architect | Edinburgh | 61 | 2011/04/25 | $320,800 | 5421 | t.nixon@datatables.net |
| Garrett | Winters | Accountant | Tokyo | 63 | 2011/07/25 | $170,750 | 8422 | g.winters@datatables.net |
| Ashton | Cox | Junior Technical Author | San Francisco | 66 | 2009/01/12 | $86,000 | 1562 | a.cox@datatables.net |
| Cedric | Kelly | Senior Javascript Developer | Edinburgh | 22 | 2012/03/29 | $433,060 | 6224 | c.kelly@datatables.net |
| Airi | Satou | Accountant | Tokyo | 33 | 2008/11/28 | $162,700 | 5407 | a.satou@datatables.net |
| Brielle | Williamson | Integration Specialist | New York | 61 | 2012/12/02 | $372,000 | 4804 | b.williamson@datatables.net |